
Sitting at the midpoint of the central dogma of gene expression, RNA holds key information for characterizing, modeling, and predicting the status of cells. However, we have not yet achieved a comprehensive understanding of gene expression regulation at the RNA level. The remaining questions are twofold, intertwined: the heterogeneity of RNA molecules and the combinatorial regulation through the arrangement of multiple regulatory elements in the linear sequence of an RNA. Understanding gene regulation at RNA level, for both endogenous and exogenous RNAs, has direct importance in guiding the design of RNA-based therapeutics.
In human cells, a single gene can often produce several isoforms of transcripts. Different isoforms may contain different RNA elements and can lead to differences in protein binding and RNA localization. Moreover, even transcripts of the same isoform can exhibit higher levels of heterogeneity such as differences in subcellular localization, diverse RNA:protein interactome, altered RNA structure, and changes in RNA modification. My lab uses integrative approaches to decipher the heterogeneity of RNA and their interactions with RNA binding proteins (RBPs). We aim to discover the principles that govern the regulation of mRNA expression through the combined effects of RNA sequence elements, RNA structure, and binding of RBPs for both endogenous and exogenous RNAs.

We developed a REMORA (RNA Encoded Molecular Recording in Adenosines), to measure the interaction between RBPs and RNAs at the single-molecule level. We engineered an improved single-stranded RNA A-to-I editor, rABE, using directed protein evolution. Fusing rABE with RBP introduces A-to-I edits adjacent to RBP binding sites on RNA, which can be identified by RNA-seq. Combined with long-read sequencing, we can identify novel isoform-specific binding patterns of RBPs. REMORA is compatible with many other RNA modification-based information encoding strategies, enabling investigation of mRNA regulation in multiple layers.
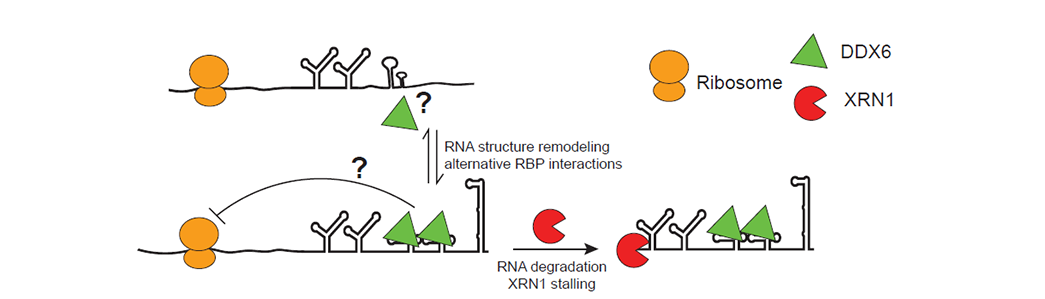
RNA structures are important cis-regulatory elements by themselves and through interactions with RBPs. Determining the in vivo structure of RNA has been a challenging task due to its heterogeneous nature. RNA structure probing methods such as SHAPE-MaP only provide an averaged measurement of all coexisting conformations of an RNA species, such measurement is further obscured by the interactions between RBPs and RNAs. We are developing experimental and computational tools to simultaneously probe RNA structure and RBP binding sites at single molecule level. This will provide rich information to decipher the heterogeneity in RBP binding and RNA structure.







Peer-reviewed manuscripts
Previews and Commentaries
Ph.D. students
We recruit graduate students through the PhD program @ SIBCB (http://www.cemcs.cas.cn/). Please email Dr. Lin (linyizhu at sibcb.ac.cn) if you are interested in internship or rotation opportunities (for incoming / first year grads).
Postdocs
We are looking for Postdocs to join us.
Qualifications:
Preferred Qualifications:
Application Process:
Interested candidates should submit the following documents to Dr. Lin (linyizhu at sibcb.ac.cn):
Please format your email title: Application-[Position]-[Name] / 应聘-[职位]-[姓名]